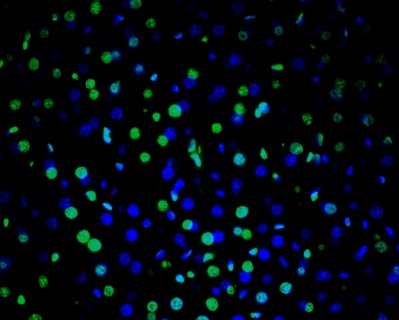
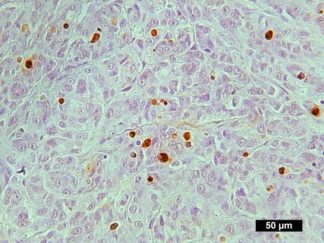
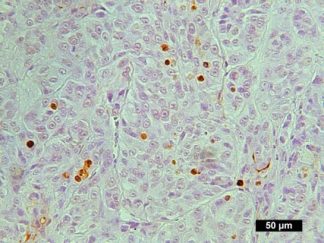
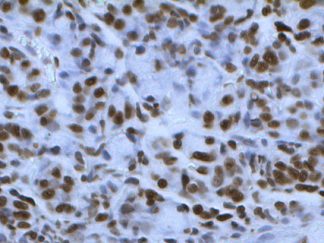
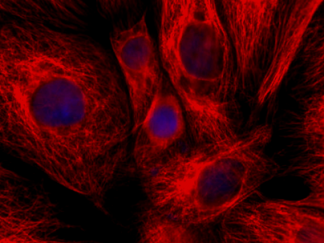
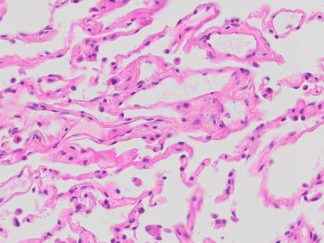
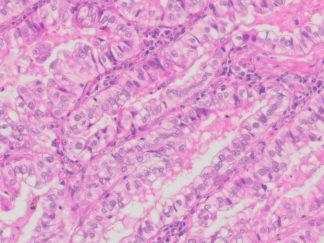
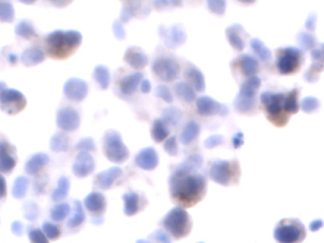
Description
DNA fragmentation represents a characteristic hallmark of apoptosis. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) is designed to detect apoptotic cells in situ that undergo extensive DNA degradation during the late stages of apoptosis. The method is based on the ability of terminal deoxynucleotidyl transferase (TdT) to label blunt ends of double-stranded DNA breaks independent of a template. In Situ TUNEL Andy488 Apoptosis Detection Kit is used for in situ apoptosis detection and visualized with green fluorescent Andy Fluor 488 under fluorescence microscope.
Kit Contents
| VB-4005G-1 | Protein K stock Solution (20×) | 0.25 ml |
| VB-4005G-2 | 1× Proteinase K working buffer | 5 ml |
| VB-4005G-3 | TdT equilibration buffer | 6 ml |
| VB-4005G-4 | TdT enzyme | 15 µl |
| VB-4005G-5 | Biotinylated dUTP | 22 µl |
| VB-4005G-6 | Streptavidin-Andy Fluor 488 | 30 µl |
| VB-4005G-7 | TUNEL positive FFPE slides | 2 slides |
Storage Condition
- Streptavidin- Andy Fluor 488: Store at 4 ºC.
- TUNEL positive FFPE slides: Store at 4 ºC.
- Others: Store at -20 ºC.
Protocol
1. Preparation of Slides
1) Frozen Sections
- Snap frozen fresh tissues in liquid nitrogen or isopentane pre-cooled in liquid nitrogen, embedded in OCT compound in cryomolds. Store the frozen tissue block at -80°C until ready for sectioning.
- Transfer the frozen tissue block to a cryotome cryostat (e.g. -20°C) prior to sectioning and allow the temperature of the frozen tissue block to equilibrate to the temperature of the cryotome cryostat.
- Section the frozen tissue block into a desired thickness (typically 5-10 µm) using the cryotome.
- Place the tissue sections onto glass slides suitable for immunohistochemistry (e.g. Superfrost)
- Sections can be stored in a sealed slide box at -80°C for later use.
- Before staining, warm slides at room temperature for 30 minutes and fix slides by 2% neutralized formalin for 30 min. Rinse slides with PBS then transfer to a Coplin jar containing ice-cold 70% ethanol for 1 h. Slides may be stored overnight in 70% ethanol at 4°C.
- Wash in PBS.
- Follow procedure for pretreatment as required with TdT reaction buffer.
2) Paraffin Sections
- Deparaffinize sections in xylene, 3×5min.
- Hydrate with 100% ethanol, 2×2min.
- Hydrate with 95% ethanol, 2×2min.
- Rinse in distilled water.
- Prepare 1ml of 1× proteinase K working solution by mixing 50µl of 20× proteinase K solution with 950 µl of 1×Proteinase K working buffer.
- Prepare carefully blot away excess water and pipette 75-100 µl of 1×proteinase K solutions to cover sections. Incubate 8~15 min at room temperature.
- Following proteinase K treatment, wash slides 3 × 5 min with ddH2
- (Optional)Inactivate endogenous peroxidases by covering sections with 2% hydrogen peroxide for 5 min at room temperature. Wash slides 3 × 5 min with ddH2O
- Follow procedure for pretreatment as required with TdT reaction buffer.
3) Adherent Cells
- Adherent cells may be cultured on glass chamber slides. Wash in PBS.
- Fix slides by 2% neutralized formalin for 15 min. Rinse slides with PBS then transfer to a Coplin jar containing ice-cold 70% ethanol for 1 h. Slides may be stored overnight in 70% ethanol at 4°C
- Wash in PBS. Inactivate endogenous peroxidases by covering sections with 2% hydrogen peroxide for 5 min at room temperature. Wash slides 3 × 5 min with ddH2
- Follow procedure for pretreatment as required with TdT reaction buffer.
4) Suspension cells
- Harvest cells and wash them twice in PBS using centrifugation (400g for 5 min) to remove residual protein.
- Adjust the cell concentration to 4–5×106 cells per mL in PBS.
- Attach cells to slides using either the adhesion or centrifugation method:
a) Adhesion method
- Clean and label the slides.
- Wash slides in PBS for 5 minutes at RT.
- Place slides in a humidified box to prevent them from drying.
- Place 20 to 50 μL of the cell suspension (at least enough to cover the well) in each well of the adhesion slides and let cells adhere at room temperature (RT) for 20 min.
- Fix slides by 2% neutralized formalin for 15 min. Rinse slides with PBS then transfer to a Coplin jar containing ice-cold 70% ethanol for 1 h. Slides may be stored overnight in 70% ethanol at 4°C .
- Inactivate endogenous peroxidases by covering sections with 2% hydrogen peroxide for 5 min at room temperature. Wash slides 3 × 5 min with ddH2
- Wash in PBS.
- Follow procedure for pretreatment as required with TdT reaction buffer.
b) Centrifugation method
- Assemble the CytoSpin centrifuge’s sample chamber, filter card, slide, and racks according to the manufacturer’s instructions.
- Load 100 μL of cells in each sample chamber.
- Centrifuge the slides at 600 rpm for 2 to 4 min.
- Remove the slides from the rack and place them on a staining rack.
- Fix slides by 2% neutralized formalin for 15 min. Rinse slides with PBS then transfer to a Coplin jar containing ice-cold 70% ethanol for 1 h. Slides may be stored overnight in 70% ethanol at 4°C
- Wash in PBS. Inactivate endogenous peroxidases by covering sections with 2% hydrogen peroxide for 5 min at room temperature. Wash slides 3 × 5 min with ddH2
- Follow procedure for pretreatment as required with TdT reaction buffer.
2. TUNEL Reaction
- Carefully blot away excess water then cover sections with TdT equilibration buffer for 10 min at room temperature.
- Preparation of TdT reaction buffer:
| 2 samples | 4 samples | 8 samples | |
| TdT equilibration buffer | 24 µl | 48 µl | 94 µl |
| TdT enzyme | 0.5 µl | 1 µl | 2.0µl |
| Biotinylated dUTP | 0.8 µl | 1.6µl | 3.2 µl |
Mix well. Prepare fresh from stock solution prior to use.
- Remove TdT equilibration buffer and cover sections with 10-12 µl of TdT reaction buffer. Incubate slides in a humidified chamber for 30 min at 37°C. In order to conserve reagents a reduced volume of TdT buffer may be carefully covered with a glass coverslip during the incubation. Take care to avoid trapping air bubbles which may lead to staining artifacts.
- Stop reaction by incubating slides 2 × 10 min in 1×SSC.
3. Visualized with Andy Fluor 488
- Rinse slides in PBS then block nonspecific binding by covering tissue sections with 2% BSA solution for 10 minutes at room temperature.
- Rinse in PBS for 3×2 minutes.
- Dilute streptavidin-Andy Fluor 488 at 1:200 in PBS to make RTU streptavidin-Andy Fluor 488.
- Detection: Incubate sections with 3-4 drops of RTU streptavidin-Andy Fluor 488 for 30 minutes at room temperature.
- Rinse in PBS for 3×2 minutes.
- Counterstain: Dilute the DAPI stock solution to 300 nM in PBS. Add approximately 300 µL of this dilute DAPI staining solution to the section/coverslip, making certain that the tissue/cells are completely covered.
- Incubate for 1–5 minutes.
- Rinse the sample several times in PBS. Drain excess buffer from the section/coverslip and mount. We recommend using a mounting medium with an antifade reagent.
- View the sample using a fluorescence microscope with appropriate filters.
Negative Control: As a negative control, omit the TdT enzyme from the TdT reaction buffer.
Note: This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
Precautions: Handle with care. Avoid contact with eyes, skin and clothing. Do not ingest. Wear gloves.